Solid-state nuclear magnetic resonance (NMR) spectroscopy can be used to detect chemical and 3D structures, as well as the dynamics of molecules and materials, by measuring the frequencies emitted by the nuclei of particular atoms subjected to radio waves in a strong magnetic field.
However, so-called chemical shift assignment is a required first step in the analysis. Each peak in the NMR spectrum is assigned to a specific atom in the molecule or substance under study. This is an extremely difficult task. Experimenting with chemical shifts can be difficult, and it usually demands time-consuming multi-dimensional correlation investigations. An alternate option would be to assign molecular solids using statistical analysis of experimental chemical shift databases, but there is no such database for molecular solids.
EPFL professors Lyndon Emsley, head of the Laboratory of Magnetic Resonance, Michele Ceriotti, head of the Laboratory of Computational Science and Modelling, and PhD student Manuel Cordova formed a team to address the issue by developing a method for assigning NMR spectra of organic crystals probabilistically, directly from their 2D chemical structures.
They began by combining the Cambridge Structural Database (CSD), a database of over 200,000 three-dimensional organic structures, with ShiftML, a machine learning algorithm they had previously developed together that allows chemical shifts to be predicted directly from the structure of molecular solids.
ShiftML was first detailed in a Nature Communications study in 2018, and it employs DFT calculations to train before making accurate predictions on new structures without the need for extra quantum computations. Though DFT precision is achieved, the approach can calculate chemical shifts for structures with fewer than 100 atoms in seconds, saving up to 10,000 times the computing cost of current DFT chemical shift computations. The method’s accuracy is independent of the size of the structure being studied, and the forecast time is proportional to the number of atoms. This paves the way for computing chemical changes in situations where it was previously impossible.
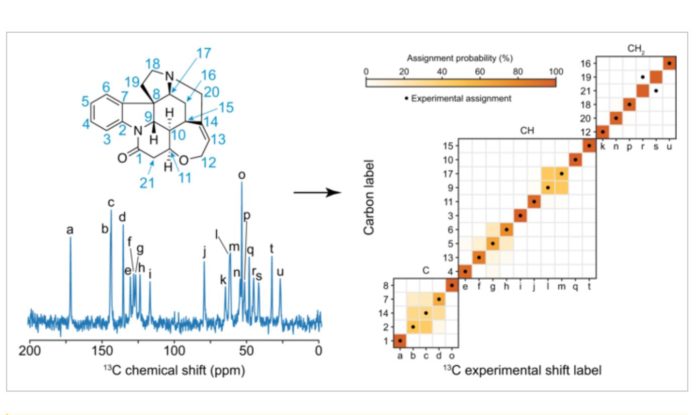
In this study, after analyzing more than 200,000 CSD compounds, they utilized ShiftML to anticipate shifts and then linked these changes to topological representations of chemical environments. Covalent bonds were represented by building a graph, which was then extended away from the molecule’s center atoms by a certain number of bonds. It was therefore possible for them to get a statistical distribution of chemical shifts for each motif by merging all of the graphs that were identical. No 3D structural features are included in this representation, which simplifies covalent bonding around the molecular atom. This allowed the researchers to use a marginalization scheme to obtain the probabilistic assignment of NMR spectra of organic crystals directly from their two-dimensional chemical structures.
In order to test their hypothesis, the researchers built a chemical shift database and then used it to predict the assignments for several organic molecules for which the carbon chemical shift assignment had already been experimentally established, including theophylline, thymol, cocaine, strychnine, AZD5718, lisinopril, ritonavir, and the K salt of penicillin G. In most cases, the two-dimensional description of the molecules yielded assignment probabilities that closely matched the empirically determined assignments.
Finally, they tested the framework’s performance against a set of 100 crystal structures containing between 10 and 20 distinct carbon atoms. The ShiftML projected shifts for each atom were utilized as the right assignment, and they were eliminated from the statistical distributions that were used to assign the molecules. In more than 80% of situations, the correct assignment was discovered among the two most likely assignments.
“This method could significantly accelerate the study of materials by NMR by streamlining one of the essential first steps of these studies,” the author concluded.
You were reading: Machine learning can help identify organic crystals in NMR spectra